Syntrichia ruralis: emerging model moss genome reveals a conserved and previously unknown regulator of desiccation in flowering plants
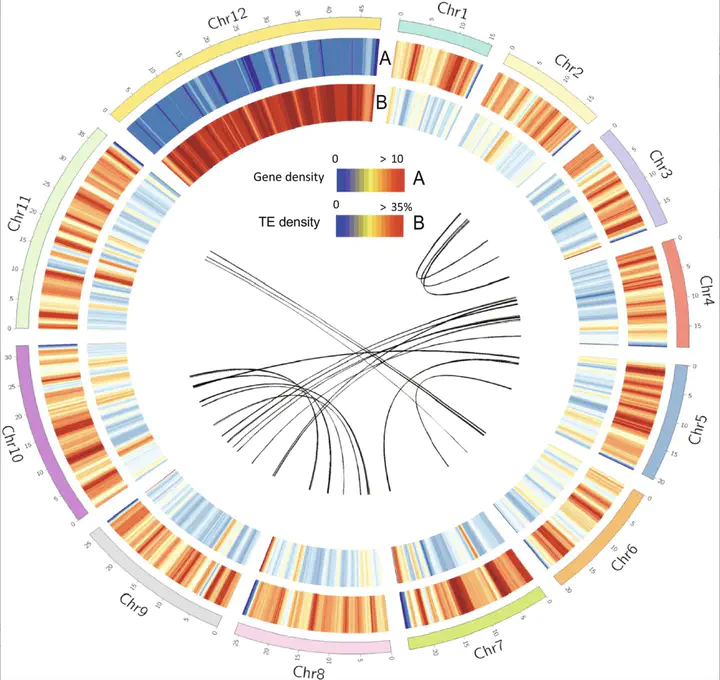 Characteristics of the Syntrichia ruralis genome. From outer to inner: The 12 chromosomes; (A) gene density in 100-kilobase (kb) genomic regions; (B) transposable element (TE) density in 100-kb genomic regions; the lines in the center reflect inter-chromosomal genomic collinearity, as defined by MCScan.
Characteristics of the Syntrichia ruralis genome. From outer to inner: The 12 chromosomes; (A) gene density in 100-kilobase (kb) genomic regions; (B) transposable element (TE) density in 100-kb genomic regions; the lines in the center reflect inter-chromosomal genomic collinearity, as defined by MCScan.Abstract
Water scarcity, resulting from climate change, poses a significant threat to ecosystems. Syntrichia ruralis, a dryland desiccation-tolerant moss, provides valuable insights into survival of water-limited conditions. We sequenced the genome of S. ruralis, conducted transcriptomic analyses, and performed comparative genomic and transcriptomic analyses with existing genomes and transcriptomes, including with the close relative S. caninervis. We took a genetic approach to characterize the role of an S. ruralis transcription factor, identified in transcriptomic analyses, in Arabidopsis thaliana. The genome was assembled into 12 chromosomes encompassing 21 169 protein-coding genes. Comparative analysis revealed copy number and transcript abundance differences in known desiccation-associated gene families, and highlighted genome-level variation among species that may reflect adaptation to different habitats. A significant number of abscisic acid (ABA)-responsive genes were found to be negatively regulated by a MYB transcription factor (MYB55) that was upstream of the S. ruralis ortholog of ABA-insensitive 3 (ABI3). We determined that this conserved MYB transcription factor, uncharacterized in Arabidopsis, acts as a negative regulator of an ABA-dependent stress response in Arabidopsis. The new genomic resources from this emerging model moss offer novel insights into how plants regulate their responses to water deprivation.